Latest vasion
The latest version is Version v2.2.0.
Example data
The Example data
Simulation data
The Simulation data
Release
Version 2.2.0 2017-10-23
1. Using two types of sequencing depths around AS events supporting inclusion and exclusion events for exon-centric strategy (see below Figure 1). Note: it is different from the previous version 2.1.0 that use RNA-seq coverage across the alternative exon (or part of exon) and the whole gene.
2. Distinction between inclusive and exclusive in the output files.
3. Reporting the loci of whole AS events instead of only reporting the loci of AS exons (or part of exons).
4. Adding another general index for assessing AS events, that is, delta_PSI (Percent Splicing In) between control and case samples.
5. Optimizing the calculation of Alternative start exons (Warning: in the case of three more continuous start exons, it might report not significant difference between samples).
6. Optimizing the calculation of MXE events.
7. Adding a new parameter "--runSepChr" . Due to some species chromosomes (e.g. Hordeum vulgare) with a huge length, the java module 'htsjdk (v2.9.0)' can hardly support to index the chromosomes, and the users can set this parameter to be False, which means CASH run without index files, but it will take more memory and more computing time.
8. Adding a new parameter "--ChrRegion" . If the users only focus on specific chromosome/regions, they can set the parameter as "--ChrRegion chr1" or set a specific region as "--ChrRegion chr1:1-9527".
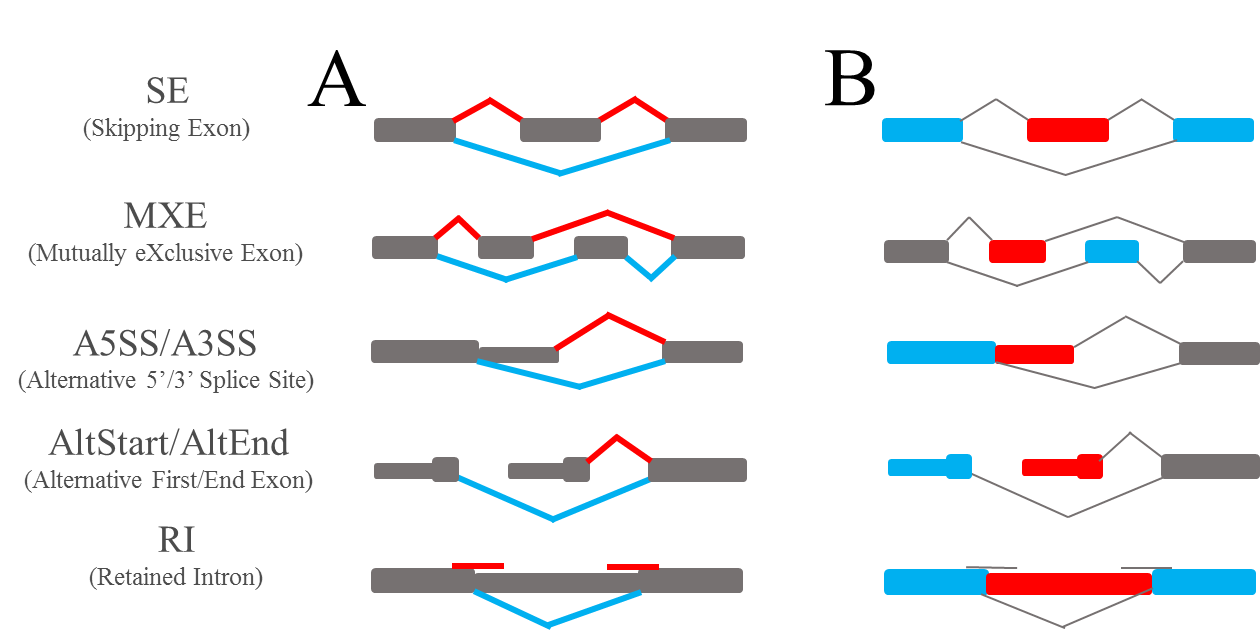
Figure 1: Detection of Alternative Splicing events. (A) Strategy I: event-centric strategy based on the expression of two types of junctions reads (i.e. inclusion and exclusion ones). The red junction reads support inclusion events and the blue ones support exclusion events. (B) Strategy II: exon-centric strategy based on the expression of two types of sequencing depths around AS exons. Sequencing depths on red exons support inclusion events and that on blue exons support exclusion events.
Thanks to Yapu Li of State Key Laboratory of Experimental Hematology and Mehrotra Sudeep of Mass. Eye and Ear for sharing experience of CASH usage and advice to improve the program.
Version 2.1.0-alpha1 2017-05-15
1. Optimized the algorithm of Pvalue calculation. In CASH, geometric weighted mean of event-centric and exon-centric strategy Pvalues is calculated to assess the differential level of AS event between samples and for some cases, the calculation likely produces falsely differentially AS events with significantly low Pvalues and to adjust the case, we added a parameter "--MergePval A/G", default value is A indicating arithmetic weighted mean and the other setting is G indicating geometric weighted mean. It is recommend to use the default value, while the results showed a poor number of differentially AS events, the users can switch A to G.
2.Add parameter "--ChrRegion" which can specify the region for CASH.
Version 2.1.0-alpha 2017-05-02
1. Optimized the memory usage.
2. Deeply optimized the chi-square test of the pvalue calculate, especially for samples more than 6vs6.
3. Discarded the function of extracting the sequences around alternative splicing sites.
Version 2.0.2 2017-04-23
1. Add parameter "--runSepChr" True/False, default is True.
In order to save the memory, cash detect splicing events for each chromosome seperately. It means cash detect one chromosome then clean the memory and detect another chromosome. So it needs index files of input bam files.
However, some species such as Hordeum vulgare, have very long chromosome and now htsjdk(v2.9.0) can hardly support the index. So inorder to run cash normally, you can set this param to False and it can run without index files, but may needs some more memory.
This bug was reported by Girma Bedada from Department of Plant Biology Uppsala BioCenter and LinneanCenter for Plant Biology. Thanks for his help.
2. Add "P-Value_Average" in output files. "P-Value_Average" is arithmetic weighted mean of event-centric strategy and exon-centric strategy Pvalues while "P-Value_Average" is geometric weighted mean of event-centric strategy and exon-centric strategy Pvalues.
It is recommend to use "P-Value_Average" as a cutoff while you want to do experiment to validate the AS site. Because small "P-Value_Average" means event-Pvalue and exon-Pvalue are both small, but small "P-Value" means one of the event-Pvalue and exon-Pvalue is small.
Version 2.0.1 2017-02-25
CASH fix some bugs in previous ASD version and importantly, this new version can reconstruct splice sites from RNA-seq data and thus provide novel alternative splicing events
that are not included in reference gene annotations(such as GTF file).