单细胞核测序技术是在单细胞水平上发展起来的技术,该技术可将冻存样本直接制核捕获,且依旧保存单细胞测序的高分辨率,能够准确区分细胞异质性,揭示单个细胞的基因结构和基因表达状态。因其对样本包容性高,核测序的出现解决了某些珍贵冻存样本,异形细胞样本无法进行单细胞实验的问题,在心肺疾病,神经发育等领域助力临床治疗和诊断,加速精准医疗时代。
1、突破性改进制核手段:跳过样本消化步骤,直接冻存组织进行制核。这种方法可以大大节省时间和资源,并且提高制核效率,细胞核捕获率高达65%-80%
2. 消除细胞解离偏差:消除由于消化偏好性造成的细胞解离偏差,保证制核质量远高于上机捕获要求,可以为后续的分析提供更准确的结果
3. 严格质量把控:烈冰全程进行严格的质量把控,从实验设计到分析产出提供一站式服务流程,确保客户获得高质量的数据和分析结果,并提供全面的支持和解决方案。
4. 核测序经验丰富,助力发表高分SCI文献
(图片来源:Jing Wu et al., Nature Neuroscience,2023)
样本类型:
1.冻存组织、培养的细胞系等,一般取黄豆粒大小;
2.-80℃冰箱保存,送样时干冰运输





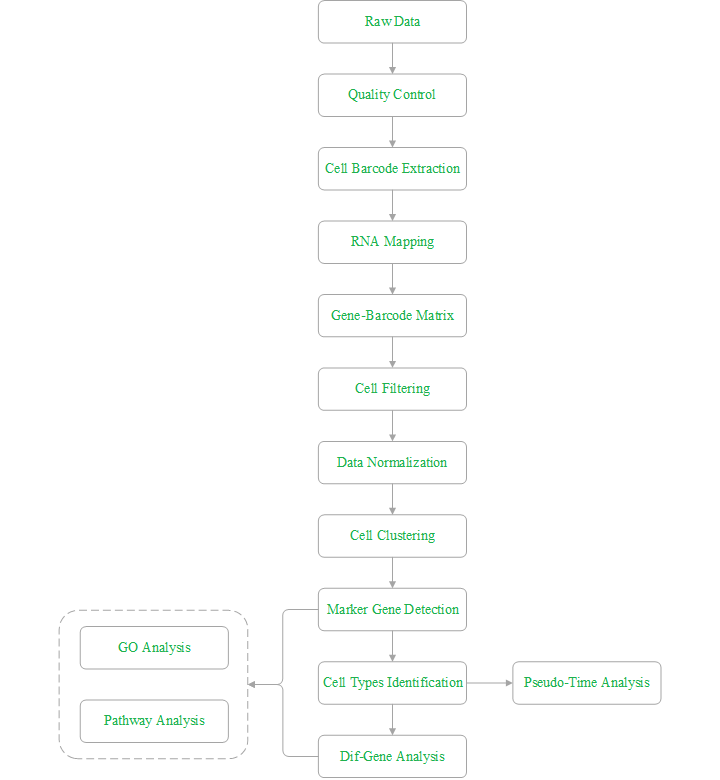
客户样本--细胞核制备--核质控检测--单细胞核捕获--细胞/转录本标签添加--文库构建--上机测序
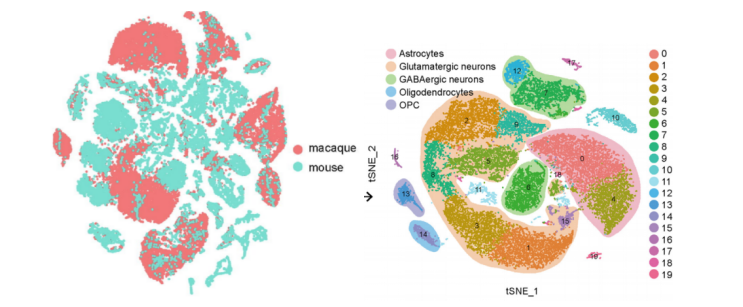
Zhang, et al. Science Bulletin, 2020 Dec.
注:图为猕猴脑组织细胞亚群鉴定t-SNE图展示
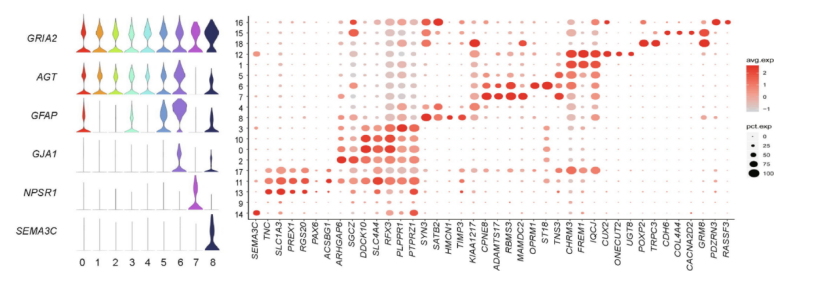
Zhang, et al. Science Bulletin, 2020 Dec.
注:Marker基因的Violin图(左)和bubble plot图(右)
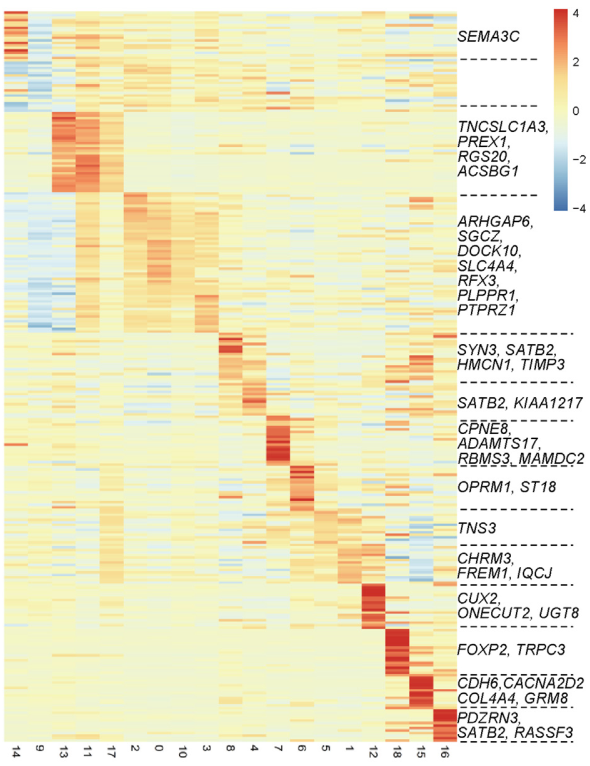
Zhang, et al. Science Bulletin, 2020 Dec.
注:该图为不同细胞亚群间差异基因聚类分析图(Heatmap)

Zhang, et al. Science Bulletin, 2020 Dec.
Zhang, et al. Science Bulletin, 2020 Dec.
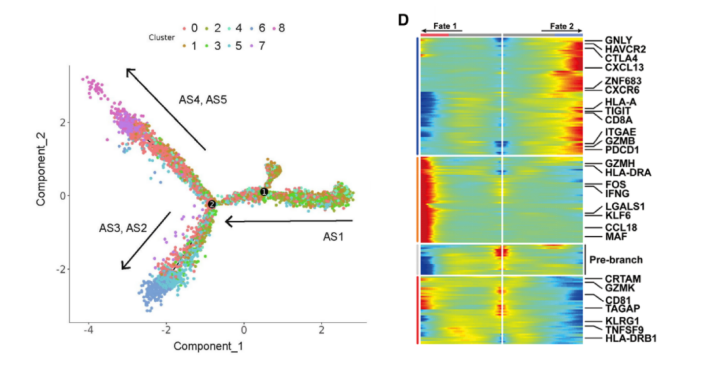
Zhang C, et al. J Immunother Cancer, 2021;9:e002312.
注:细胞间状态转换的pesudotime轨迹图(左)和Heatmap图(右)
烈冰助力文献:
[1] Single-nucleus transcriptomic mapping of blast-induced traumatic brain injury in mice hippocampus,Scientific Data,2023,IF=9.8
[2] Integrating spatial and single-nucleus transcriptomic data elucidates microglial-specific responses in female cynomolgus macaques with depressive-like behaviors,Nature Neuroscience,2023, IF=25
[3] Targeting gut microbiota–derived kynurenine to predict and protect the remodeling of the pressure_overloaded young heart,Science Advances,2023,IF=13.6
[4] Silencing of NOTCH3 Signaling in Meniscus Smooth Muscle Cells Inhibits Fibrosis and Exacerbates Degeneration in a HEYL-Dependent Manner, Advanced Science, 2023,IF=15.1
[5] The molecular taxonomy of primate amygdala via single-nucleus RNA-sequencing analysis,Science Bulletin,202012, IF=18.9